Groundbreaking molecular profiling method reveals genes associated with the vulnerability to Alzheimer’s disease
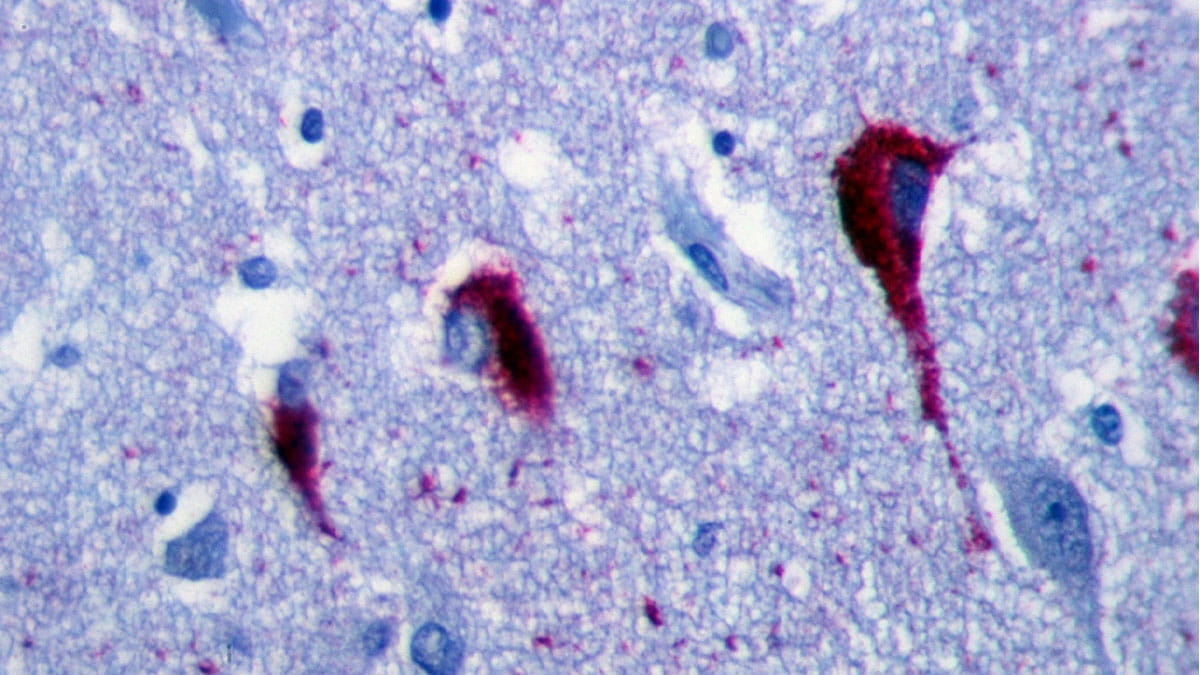 Alzheimer’s disease (AD) is the most common form of dementia, affecting an estimated 6.5 million Americans over the age of 65. AD is characterized by degeneration of neural connections in the brain, causing symptoms such as memory loss, mental decline and depression.
Alzheimer’s disease (AD) is the most common form of dementia, affecting an estimated 6.5 million Americans over the age of 65. AD is characterized by degeneration of neural connections in the brain, causing symptoms such as memory loss, mental decline and depression.
“Understanding why some neurons in particular brain regions are highly vulnerable to degeneration is of fundamental importance for neurodegenerative diseases, including AD,” says Harry Fu, PhD, assistant professor of Neuroscience at The Ohio State University College of Medicine.
Dr. Fu worked with Qin Ma, PhD, associate professor of Biomedical Informatics at the Ohio State College of Medicine, and a group of collaborators to identify the molecular mechanisms underlying the cellular and regional vulnerability in AD and AD-related dementias. They are hoping to develop new therapeutics targeting identified molecular determinants for these diseases for which there are no cures.
Their latest publication, “Spatially resolved transcriptomics reveals genes associated with the vulnerability of middle temporal gyrus in Alzheimer’s disease,” recently published in Acta Neuropathologica Communications, is the first to characterize human postmortem brains with AD using 10x Genomics’ Visium Spatial Gene Expression resolved transcriptomics.
“This study builds on our previously published research about how a tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology,” Dr. Fu says. “The technique of spatially resolved transcriptomics such as 10x Visium platform is a groundbreaking molecular profiling method that allows scientists to characterize complex transcriptional landscapes within a spatial context, which is essential for the understanding of biology and complex disease.”
Using 10x Visium in combination with immunofluorescent staining of Alzheimer’s neuropathology (i.e., amyloid beta and tau protein aggregates), Dr. Fu was able to compare transcriptomic profiles between AD and control subjects without the loss of spatial information. “This approach generated the first-of-its-kind Visium data for human AD and enables a better understanding of molecular mechanisms underlying the region-specific distribution AD neuropathology within the spatial context," Dr. Ma says.
Spatial capture technology powers the Visium Spatial platform using spatially barcoded mRNA-binding oligonucleotides. Each spot on the Visium platform has a unique spatial barcode and thus locates the position of the brain tissue mounted on the Visium gene expression slide.
“Further, we can determine the specific area of the brain with AD neuropathology by immunostaining the gene expression brain section or adjacent brain section with antibodies specific against amyloid beta and tau protein aggregates,” Dr. Fu says.
In addition, fresh frozen tissue is permeabilized to release mRNA from the cells, which binds to the spatially barcoded oligonucleotides present on the spots. A reverse transcription reaction produces cDNA from the captured mRNA. The barcoded cDNA is then pooled for downstream processing to generate a sequencing-ready library.
Meanwhile, with bioinformatics tools and computational analyses, Dr. Fu was able to determine the molecular make-up of AD and control spots as well as within each AD brain tissue, spots with AD neuropathology versus spots without AD neuropathology. The alterations of gene signatures in certain spots may contribute to the regional vulnerability in AD.
Overall, the Visium analysis highlights the molecular composition of the anatomical architecture of the human middle temporal gyrus and the complexity of glial-neuronal hub gene interactions in response to a range of AD-associated neuropathology, which may contribute to the cellular and regional vulnerability at early stages of AD.
If the gene signatures that were identified and validated in Dr. Fu’s study can be repeated by large population studies, they can be used as biomarkers for the diagnosis and prognosis of patients with AD. These results can also provide insights for developing therapeutics for prevention and treatment of AD and AD-related dementias.
The study was mainly supported by the National Institutes of Health, National Institute of General Medical Sciences, Alzheimer’s Association, the U.S. Department of Defense, and the 10x Genomics 2021 Neuroscience Challenge award.
Explore more stories of innovation, clinical care, education and community outreach at The Ohio State University College of Medicine.



